SSPACE SCAFFOLDING FREE DOWNLOAD
It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide. Hi all, I have used Minia to assemble a contigs set from my paired end reads, as Minia instructe I have multiple libraries supposedly made at insert sizes from 6kb to 20kb. We demonstrate that applying contig extension prior to scaffolding can further enhance the outcomes. Optionally, the pre-assembled contigs can be extended using sequence reads that could not be mapped. Related articles in Web of Science Google Scholar. SSPACE is able to scaffold large genomes in a reasonable amount of time even when using huge datasets. 
| Uploader: | Akinokinos |
| Date Added: | 18 September 2010 |
| File Size: | 42.81 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 73165 |
| Price: | Free* [*Free Regsitration Required] |
This feature is especially designed to incorporate the paired-read datasets that were not used in the pre-assembly. BioinformaticsVolume 27, Issue 4, 15 FebruaryPages —, https: Our method is suited both for prokaryotic and eukaryotic genomes and in comparison with two built-in scaffolders SSPACE achieves competitive results. Extension of scaffolds is aborted if scaffoleing a contig has no links with other contigs or the ratio for alternatives is exceeded.
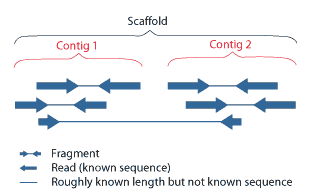
Scaffolding pre-assembled contigs using SSPACE.
Email alerts New issue alert. De novo assembly tools play a main role in reconstructing genomes from next-generation sequencing NGS data and usually yield a number of contigs. The percentage of shared nucleotides is calculated from the number of nucleotides that are involved in contig-pairs.
Supplementary data are available at Bioinformatics online. We demonstrate that applying contig extension prior to scaffolding can further enhance the outcomes. Hereafter a post-filtering step is applied to remove duplicate read-pairs.
Although the latter process is a crucial scaffoldng in finishing genomes, scaffolding algorithms are often built-in functions in de novo assembly tools and cannot be independently controlled. Nonetheless, the majority of assembly tools provide a scaffolding option only as a built-in function which cannot be independently controlled, among which SSAKE Warren et al. The remaining read pairs are mapped against the pre-assembled contigs using Bowtie Langmead et al.
I am using SOAPdenovo. Marten Boetzer, Christiaan V. Sdpace is able to scaffold large genomes in a reasonable amount of time even when using huge datasets.
Scaffolding pre-assembled contigs using SSPACE | Bioinformatics | Oxford Academic
Otherwise a ratio is calculated between the two best alternatives. All analyses were performed scaffoldingg a 32 GB Linux machine. Looking for your next opportunity? The minimum number of links required for matching unambiguous contig links was set to 5. This can partly be attributed to the presence of repetitive elements on the genome. I used the following command to estimate insert size using bbmap:.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
Oxford University Press is a department of the University of Oxford. For both genomes, all sequence reads were assembled ssace single-ends with Abyss and subsequently scaffolded into supercontigs using the Abyss and SSPACE scaffolders. Both methods significantly reduce the number of contigs in a comparable manner, although SSPACE achieves overall better results.
Scaffolding pre-assembled contigs using SSPACE.
I want to use a paired end library for scaffolding my contigs. According to the manual: I have been trying to assemble a nematode genome denovo from scarfolding 6 months.
Our program has been designed to allow for multiple library input sets that are scaffolded in a hierarchical manner starting with small insert libraries.
The task is to puzzle back millions of short sequence reads into a limited set of contiguous sequences contigsalthough in practice the total number can be rather high. It can also be observed that the scaffolding of contig-extension prior to scaffolding yields slightly less scaffolds.
Hi, I have a weird situation. In brief, putative contig pairs pre-scaffolding stage are computed based on the position of the paired reads on different contigs. Analyses on the giant panda dataset were performed similarly to the previous two sets. Optionally, the pre-assembled contigs can be extended using sequence reads that could not be mapped.


Comments
Post a Comment